Introduction to Random Strings(무작위 문자열)
https://rosalind.info/problems/prob/
ROSALIND | Introduction to Random Strings
It appears that your browser has JavaScript disabled. Rosalind requires your browser to be JavaScript enabled. Introduction to Random Strings solved by 4930 2012년 12월 4일 7:03:37 오전 by tvinar Topics: Probability Modeling Random Genomes We already
rosalind.info
Problem
An array is a structure containing an ordered collection of objects (numbers, strings, other arrays, etc.). We let A[k]A[k] denote the kk-th value in array AA. You may like to think of an array as simply a matrix having only one row.
A random string is constructed so that the probability of choosing each subsequent symbol is based on a fixed underlying symbol frequency.
GC-content offers us natural symbol frequencies for constructing random DNA strings. If the GC-content is xx, then we set the symbol frequencies of C and G equal to x2x2 and the symbol frequencies of A and T equal to 1−x21−x2. For example, if the GC-content is 40%, then as we construct the string, the next symbol is 'G'/'C' with probability 0.2, and the next symbol is 'A'/'T' with probability 0.3.
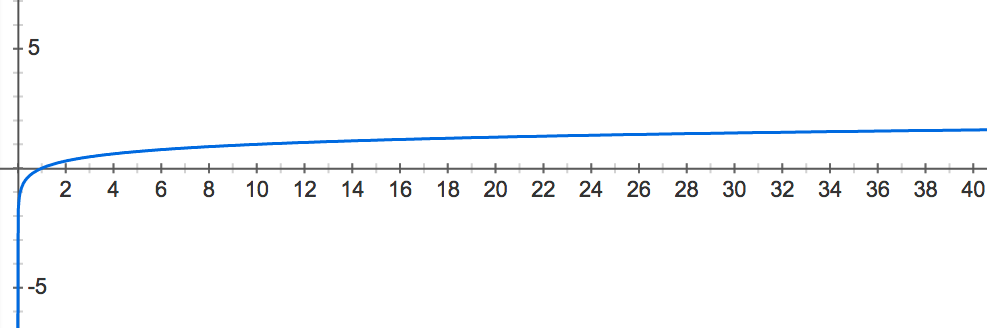
In practice, many probabilities wind up being very small. In order to work with small probabilities, we may plug them into a function that "blows them up" for the sake of comparison. Specifically, the common logarithm of xx (defined for x>0x>0 and denoted log10(x)log10(x)) is the exponent to which we must raise 10 to obtain xx.
See Figure 1 for a graph of the common logarithm function y=log10(x)y=log10(x). In this graph, we can see that the logarithm of xx-values between 0 and 1 always winds up mapping to yy-values between −∞−∞ and 0: xx-values near 0 have logarithms close to −∞−∞, and xx-values close to 1 have logarithms close to 00. Thus, we will select the common logarithm as our function to "blow up" small probability values for comparison.
Given: A DNA string ss of length at most 100 bp and an array AA containing at most 20 numbers between 0 and 1.
Return: An array BB having the same length as AA in which B[k]B[k] represents the common logarithm of the probability that a random string constructed with the GC-content found in A[k]A[k] will match ss exactly.
Sample Dataset
ACGATACAA
0.129 0.287 0.423 0.476 0.641 0.742 0.783
Sample Output
-5.737 -5.217 -5.263 -5.360 -5.958 -6.628 -7.009
이 문제는 1개의 문자열이 주어지고 그 다음 A배열이 주어진다. A배열은 각각 GC 비율을 의미한다. B배열 k번째 인덱스는 A배열의 k번째 인덱스이 GC비율과 연관이 있다. GC비율에 따라 무작위 문자열을 만들었을때, 이 문자열이 주어진 문자열과 완전히 같은 확률이 B배열의 인자이다. 이 확률이 너무나도 작기에 상용로그를 씌워서 넣는다
import math
if __name__=='__main__':#직접 실행시
with open(r'파일경로','r') as file:
string=next(file).rstrip()#주어진 문자열
A=list(map(float,(next(file).rstrip()).split()))#A배열
B=[]
for i in A:#i가 GC비율
p=1
pA,pC=(1-i)/2,i/2#A/T확률, G/C확률
for s in string:
if s=='A' or s=='T':
p*=pA
elif s=='C' or s=='G':
p*=pC
B.append(round(math.log10(p),3))#상용로그 씌우고 소수점3자리까지
print(*B)